What is Starsim?
Starsim is a framework for modeling the spread of diseases among agents via dynamic transmission networks. Starsim supports:
- Co-transmission of multiple diseases at once, capturing how they interact biologically and behaviorally
- Non-infectious diseases, either on their own or as factors affecting the transmission or mortality of infectious diseases
- Detailed modeling of mother-child relationships starting from conception, allowing investigation of infant and childhood diseases
- Multiple types of transmission network, including theoretical (e.g. Erdős–Rényi) and realistic (e.g. age-assortative sexual partnerships)
- Different intervention types, such as vaccines or treatments, and showing their impact through different delivery methods such as mass campaigns or targeted outreach
- Automated calibration to data, plus careful handling of random numbers to minimize variance between simulations
- AI-accelerated development via our dedicated MCP server that you can use with your favorite code editor
Starsim is available for both Python and R, and is fully open-source under the MIT license.
Why Starsim?
High performance
Array computations and just-in-time compilation mean Starsim achieves C++ speeds from pure Python. Starsim runs on laptops, not supercomputers, via either R or Python.
Easy to use
Starsim's modular structure means you can reuse or adapt existing disease models, transmission networks, and demographics. Mix, match, and modify any module you want.
Global community
Starsim is a community, not a product. We believe that diversity, transparency, and collaboration are essential for achieving real-world health outcomes.
Installation
If you have Python, you can install Starsim:
> pip install starsim
Or from R:
devtools::install_github("starsimhub/rstarsim")
library(starsim)
init_starsim()Examples
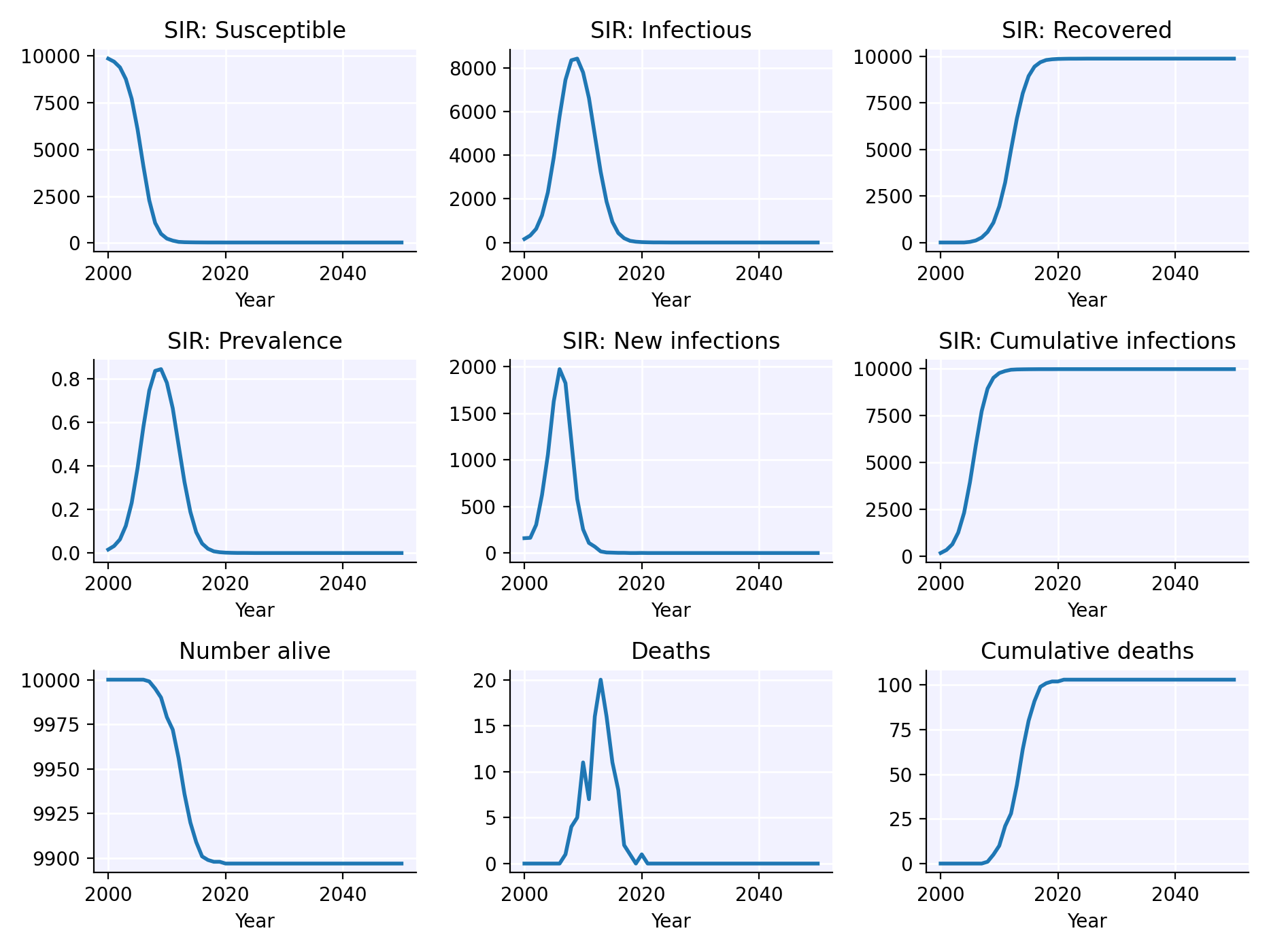
This is what an extremely simple Starsim simulation looks like:
- Create a susceptible-infectious-recovered (SIR) disease model with default parameters.
- Create a random transmission network between agents (also with default parameters).
- Run the simulation and plot the results.
import starsim as ss
sim = ss.Sim(diseases='sir', networks='random') # Create the sim
sim.run() # Run the sim
sim.plot() # Plot the results
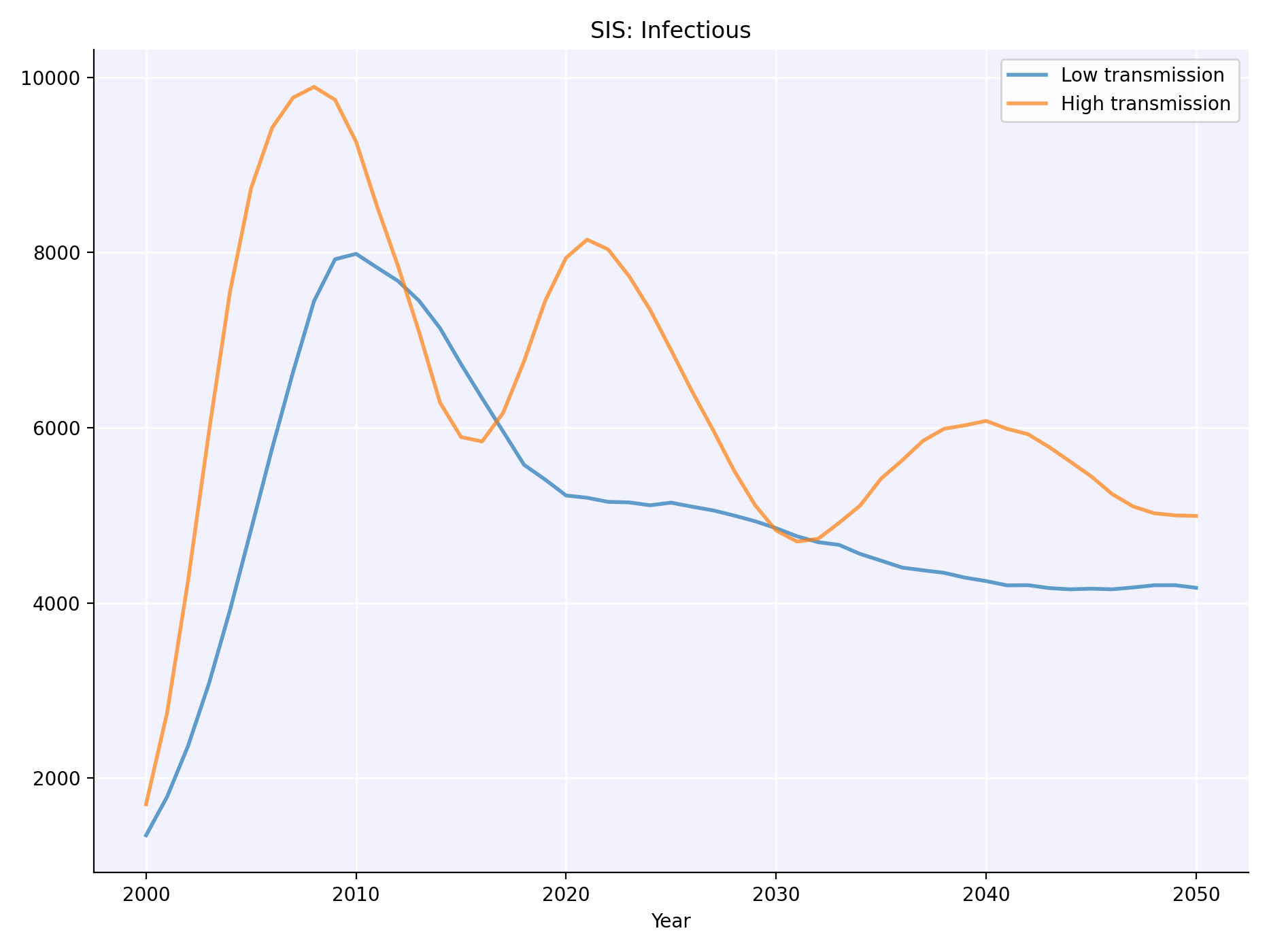
You can easily customize model parameters, and run simulations in parallel:
- Create a dictionary defining the parameters of the simulation.
- Modify only those parameters you want to differ between scenarios.
- Run the simulations in parallel, and plot the results you are interested in.
import starsim as ss
import sciris as sc
# Set the parameters for the baseline simulation
pars1 = sc.objdict( # Note: can also use regular Python dictionary
n_agents = 10_000, # Number of agents to simulate
networks = sc.objdict( # *Networks* add detail on how the agents interact with each other
type = 'random', # Here, we use a 'random' network
n_contacts = 4 # Each person has an average of 4 contacts with other people
),
diseases = sc.objdict( # *Diseases* add detail on what diseases to model
type = 'sis', # Here, we're creating an SIS disease
init_prev = 0.1, # Proportion of the population initially infected
beta = 0.1, # Probability of transmission between contacts
)
)
# Make a modified version of the parameters for the scenario
pars2 = pars1.copy(deep=True)
pars2.diseases.beta = 0.2
# Create the simulations
s1 = ss.Sim(pars1, label='Low transmission')
s2 = ss.Sim(pars2, label='High transmission')
# Run and plot the simulations
msim = ss.parallel(s1, s2)
msim.plot('sis_n_infected')
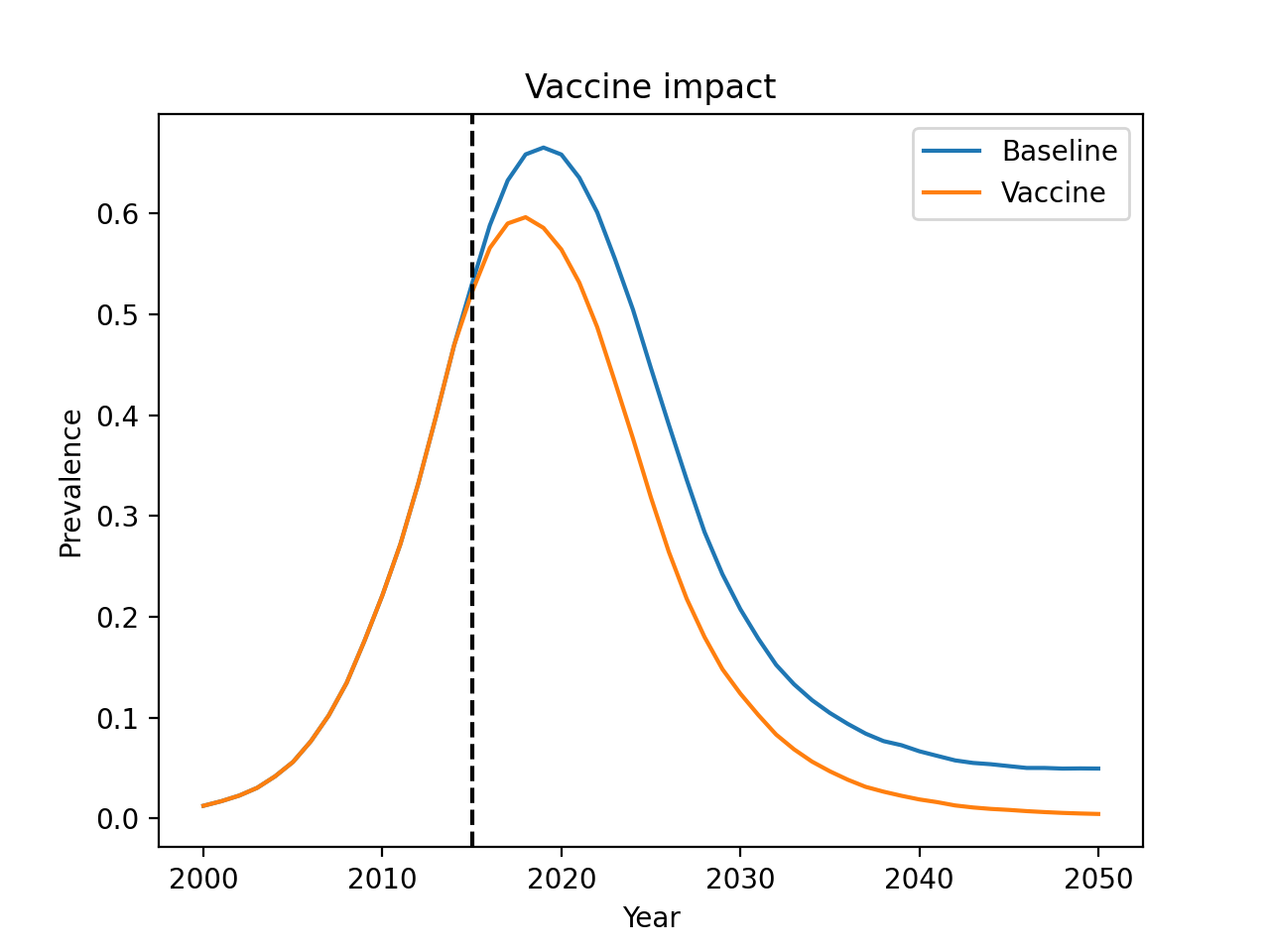
Everything in Starsim can be customized, including diseases, demographics, and intervention.
This example shows how to write custom interventions, namely a vaccine product and vaccination campaign:
import starsim as ss
import matplotlib.pyplot as plt
# Define the simulation parameters
pars = dict(
n_agents = 20_000,
birth_rate = 20,
death_rate = 15,
networks = dict(
type = 'random',
n_contacts = 4
),
diseases = dict(
type = 'sir',
dur_inf = 10,
beta = 0.1,
)
)
# Create the product: a vaccine with 50% efficacy
my_vaccine = ss.sir_vaccine(efficacy=0.5)
# Create the vaccine campaign
campaign = ss.routine_vx(
start_year = 2015, # Begin vaccination in 2015
prob = 0.2, # 20% coverage
product = my_vaccine # Use the MyVaccine product
)
# Now create two sims: a baseline sim and one with the intervention
sim_base = ss.Sim(pars=pars)
sim_intv = ss.Sim(pars=pars, interventions=campaign)
# Run sims in parallel
sims = ss.parallel(sim_base, sim_intv).sims
base = sims[0].results
vax = sims[1].results
# Plot
plt.figure()
plt.plot(base.yearvec, base.sir.prevalence, label='Baseline')
plt.plot(vax.yearvec, vax.sir.prevalence, label='Vaccine')
plt.axvline(x=2015, color='k', ls='--')
plt.title('Vaccine impact')
plt.xlabel('Year')
plt.ylabel('Prevalence')
plt.legend()
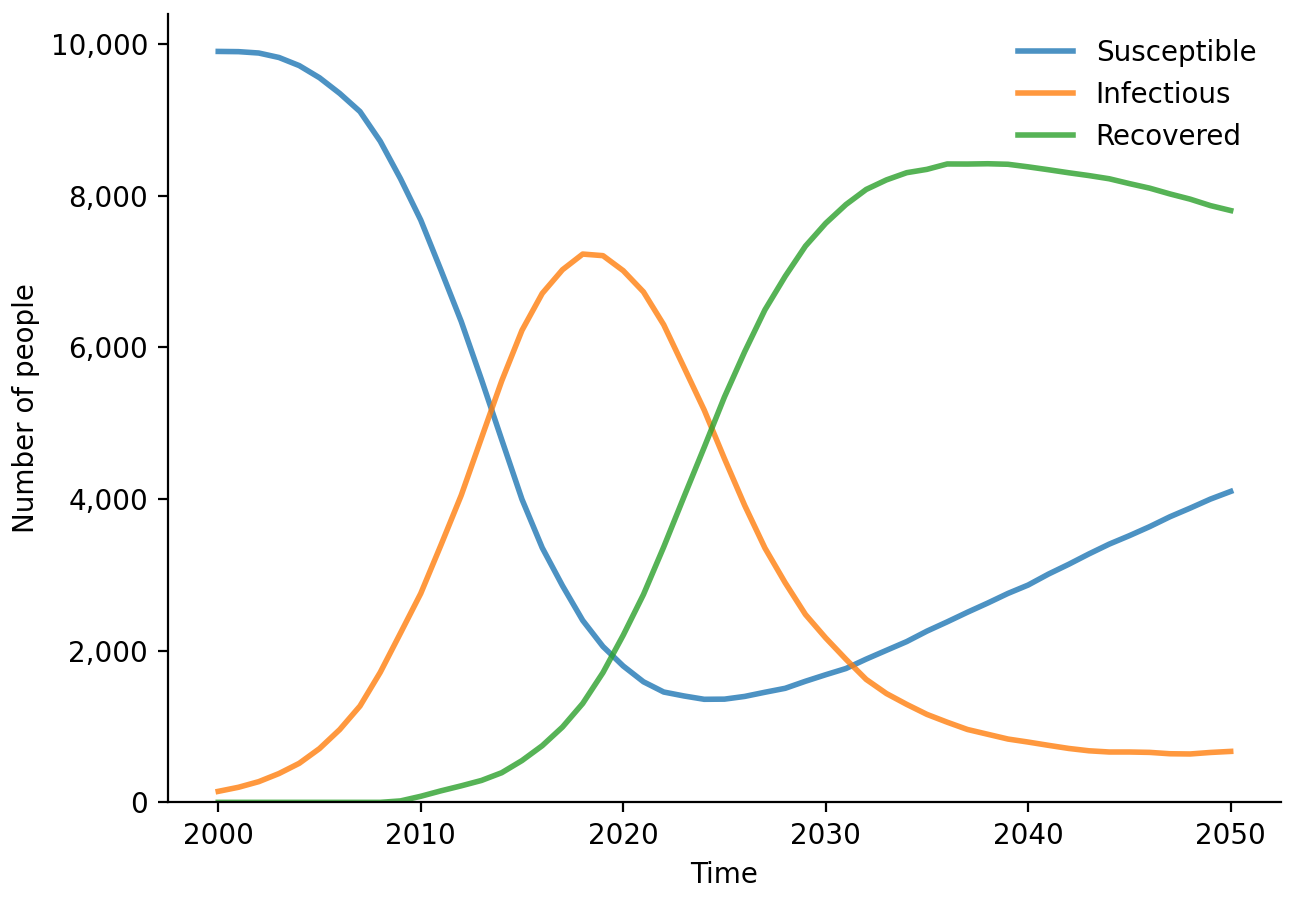
Starsim can be run from R just as easily as from Python:
# Load Starsim
library(starsim)
load_starsim()
# Set the simulation parameters
pars <- list(
n_agents = 10000,
birth_rate = 20,
death_rate = 15,
networks = list(
type = 'randomnet',
n_contacts = 4
),
diseases = list(
type = 'sir',
dur_inf = 10,
beta = 0.1
)
)
# Create, run, and plot the simulation
sim <- ss$Sim(pars)
sim$run()
sim$diseases$sir$plot()
Models
The Starsim ecosystem currently consists of the following models:
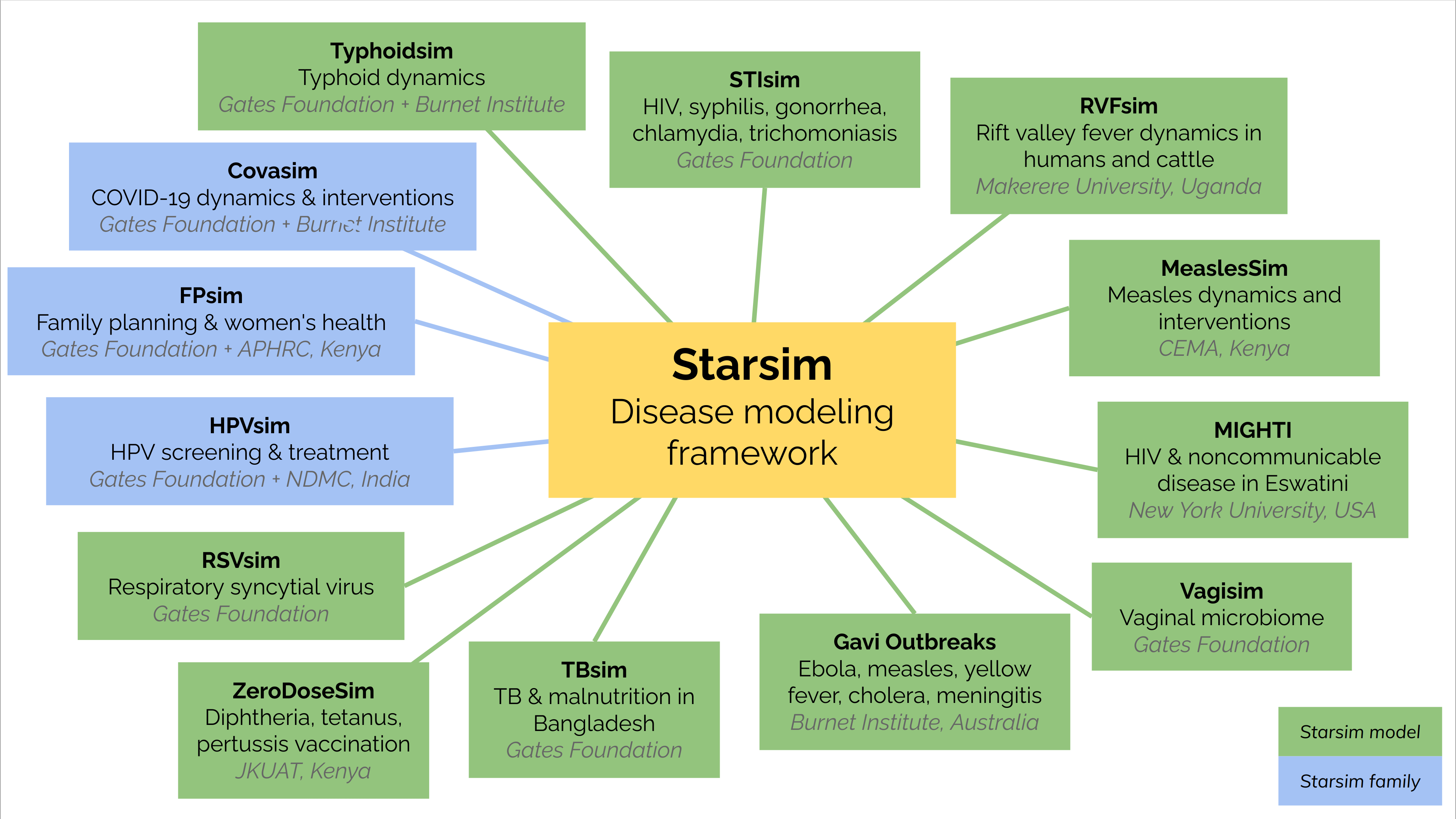
Since the Starsim framework was only released in mid 2024, most of these models are still under development and are
not yet available for public use. However, STIsim is available on GitHub,
and Gavi Outbreaks has been published.
Other models based on Starsim principles (Covasim,
HPVsim, and FPsim) are also available.
Events
Upcoming events
Nov. 30-Dec. 3, 2025: San Diego, USA
Past events
Nov. 18, 2024: Silver Spring, USA
Nov. 8, 2024: Bangkok, Thailand
Oct. 3, 2024: Seattle, USA
Jul. 25, 2024: Munich, Germany
Jul. 10, 2024: Tacoma, USA
Apr. 8-19, 2024: Nairobi, Kenya
Paper
Starsim has not yet been published. But if you want to cite it, please use:
Cliff Kerr, Robyn Stuart, Romesh Abeysuriya, Jamie Cohen, Paula Sanz-Leon, Alina Muellenmeister, Daniel Klein (2024). Starsim: A fast, flexible toolbox for agent-based modeling of health and disease. In preparation.
