Running TB HIV scenarios¶
Usage sample (with scenarios and plotting)
To use the hiv comorbidity features, you will need to add these 3 things to the simulation:
HIV diseaseto the disease list: Configure it as requested. Also, please note that if no intervention is specified, the model will use a initial prevalence and art coverage as specified in the HIV disease.TB_HIV connectorto the connector list: You will have the hability to update the values of each modifier as specified in the connector.HivInterventionto the intervention list: The same intervention can be used for managing the art coverage and the prevalence depending on the specified parameters.
[6]:
# Import required packages
import matplotlib.pyplot as plt
import numpy as np
import sciris as sc
import tbsim as mtb
import starsim as ss
Build TB-HIV Simulation¶
[7]:
def build_tbhiv_sim(simpars=None, tbpars=None, hivinv_pars=None) -> ss.Sim:
"""Build a TB-HIV simulation with current disease and intervention models."""
# Set up the simulation parameters:
default_simpars = dict(
unit='day', dt=7,
start=ss.date('1980-01-01'), stop=ss.date('2025-12-31'),
rand_seed=123,
verbose=0,
)
if simpars: default_simpars.update(simpars)
# People:
n_agents = 1000
extra_states = [ss.FloatArr('SES', default=ss.bernoulli(p=0.3))]
people = ss.People(n_agents=n_agents, extra_states=extra_states)
# Disease 1: TB:
pars = dict(beta=ss.beta(0.01), init_prev=ss.bernoulli(p=0.25), rel_sus_latentslow=0.1)
if tbpars:
pars.update(tbpars)
tb = mtb.TB(pars=pars)
# Disease 2: HIV:
hiv_pars = dict(init_prev=ss.bernoulli(p=0.10), init_onart=ss.bernoulli(p=0.50))
hiv = mtb.HIV(pars=hiv_pars)
# Network:
network = ss.RandomNet(pars=dict(n_contacts=ss.poisson(lam=2), dur=0))
# Connector: TB-HIV
connector = mtb.TB_HIV_Connector()
# Interventions: HivInterventions
interventions = []
if hivinv_pars is not None:
hiv.update_pars(hiv_pars)
hivinv_pars = hivinv_pars or dict(
mode='both',
prevalence=0.20,
percent_on_ART=0.20,
minimum_age=15,
max_age=49,
start=ss.date('2000-01-01'), stop=ss.date('2010-12-31'),
)
hiv_intervention = mtb.HivInterventions(pars=hivinv_pars)
interventions = [hiv_intervention]
# Create the simulation:
sim = ss.Sim(
people=people,
diseases=[tb, hiv],
interventions=interventions,
networks=network,
connectors=[connector],
pars=default_simpars,
)
return sim
Run HIV Intervention Scenarios¶
This function runs multiple intervention strategies, each with varying HIV prevalence, ART coverage, and target age ranges.
[11]:
def run_scenarios():
"""Run the scenarios and return the results."""
scenarios = {
'baseline': None,
'early_low_delivery_both': dict(mode= 'both', prevalence=0.10, percent_on_ART=0.10,
minimum_age=15, max_age=49,
start=ss.date('1990-01-01'), stop=ss.date('2000-12-31')),
'high_coverage_delivery_both': dict(mode= 'both', prevalence=0.25, percent_on_ART=0.75,
minimum_age=10, max_age=60,
start=ss.date('2000-01-01'), stop=ss.date('2025-12-31')),
'infection_1990_delivery': dict(mode= 'infection', prevalence=0.10,
minimum_age=15, max_age=49,
start=ss.date('1990-01-01'), stop=ss.date('2000-12-31')),
'infection_2000_delivery': dict(mode= 'infection', prevalence=0.25,
minimum_age=10, max_age=60,
start=ss.date('2000-01-01'), stop=ss.date('2025-12-31')),
'early_art_delivery': dict(mode= 'art', percent_on_ART=0.10,
minimum_age=15, max_age=49,
start=ss.date('1990-01-01'), stop=ss.date('2025-12-31')),
'high_art_delivery': dict(mode= 'art', percent_on_ART=0.75,
minimum_age=10, max_age=60,
start=ss.date('2000-01-01'), stop=ss.date('2025-12-31')),
}
flat_results = {}
for name, hivinv_pars in scenarios.items():
print(f'Running scenario: {name}')
sim = build_tbhiv_sim(hivinv_pars=hivinv_pars)
sim.run()
flat_results[name] = sim.results.flatten()
return flat_results
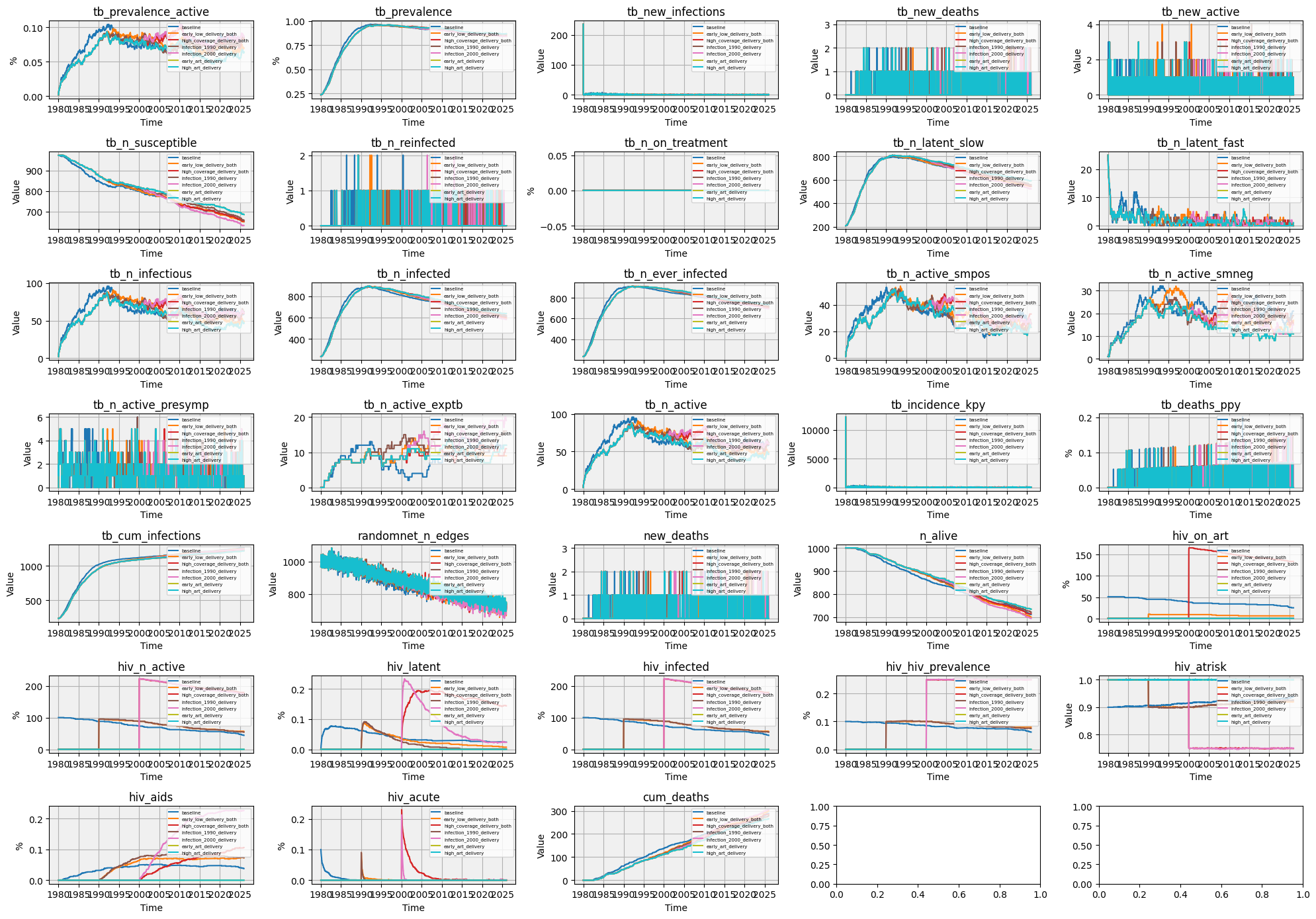
Plotting Results¶
We extract relevant metrics across scenarios and generate time series plots to visualize their trends. Metrics can be filtered by keywords or exclusions.
[12]:
def plot_results(flat_results, keywords=None, exclude=['15']):
metrics = sorted({k for flat in flat_results.values() for k in flat.keys() if (not keywords or any(kw in k for kw in keywords))}, reverse=True)
metrics = [m for m in metrics if not any(excl in m for excl in exclude)]
n_metrics = len(metrics)
if n_metrics > 0:
n_cols = 5
n_rows = int(np.ceil(n_metrics / n_cols))
fig, axs = plt.subplots(n_rows, n_cols, figsize=(20, n_rows*2))
axs = axs.flatten()
cmap = plt.cm.get_cmap('tab10', len(flat_results))
for i, metric in enumerate(metrics):
ax = axs[i]
for j, (scenario, flat) in enumerate(flat_results.items()):
if metric in flat:
result = flat[metric]
ax.plot(result.timevec, result.values, label=scenario, color=cmap(j))
ax.set_title(metric)
ax.set_ylabel('%' if max(result.values) < 1 else 'Value')
ax.set_xlabel('Time')
ax.grid(True)
ax.legend(loc='upper right', fontsize=6 if len(flat_results) <= 5 else 5)
ax.set_facecolor('#f0f0f0')
plt.tight_layout()
plt.savefig(f'{sc.thisdir()}/tbhiv_scenarios.png', dpi=300)
plt.show()
Plotting¶
[13]:
# Run everything
flat_results = run_scenarios()
plot_results(flat_results)
Running scenario: baseline
Running scenario: early_low_delivery_both
HIV intervention present, skipping initialization.
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:112: RuntimeWarning:
Not enough acute cases to revert. Expected: 1, Available: 0
ss.warn(msg=f"Not enough acute cases to revert. Expected: {-delta}, Available: {len(acute_uids)}")
Running scenario: high_coverage_delivery_both
HIV intervention present, skipping initialization.
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:112: RuntimeWarning:
Not enough acute cases to revert. Expected: 1, Available: 0
ss.warn(msg=f"Not enough acute cases to revert. Expected: {-delta}, Available: {len(acute_uids)}")
Running scenario: infection_1990_delivery
HIV intervention present, skipping initialization.
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:112: RuntimeWarning:
Not enough acute cases to revert. Expected: 1, Available: 0
ss.warn(msg=f"Not enough acute cases to revert. Expected: {-delta}, Available: {len(acute_uids)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:112: RuntimeWarning:
Not enough acute cases to revert. Expected: 2, Available: 0
ss.warn(msg=f"Not enough acute cases to revert. Expected: {-delta}, Available: {len(acute_uids)}")
Running scenario: infection_2000_delivery
HIV intervention present, skipping initialization.
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:112: RuntimeWarning:
Not enough acute cases to revert. Expected: 1, Available: 0
ss.warn(msg=f"Not enough acute cases to revert. Expected: {-delta}, Available: {len(acute_uids)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:112: RuntimeWarning:
Not enough acute cases to revert. Expected: 2, Available: 0
ss.warn(msg=f"Not enough acute cases to revert. Expected: {-delta}, Available: {len(acute_uids)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:112: RuntimeWarning:
Not enough acute cases to revert. Expected: 3, Available: 0
ss.warn(msg=f"Not enough acute cases to revert. Expected: {-delta}, Available: {len(acute_uids)}")
Running scenario: early_art_delivery
HIV intervention present, skipping initialization.
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 19, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 18, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 17, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 16, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 15, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
Running scenario: high_art_delivery
HIV intervention present, skipping initialization.
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 133, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 132, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 131, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 130, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 129, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 128, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 127, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 126, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 125, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 124, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 123, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 122, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 121, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 120, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 119, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 118, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 117, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 116, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 115, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 114, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 113, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 112, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 111, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/Users/mine/git/hivtb/tbsim/comorbidities/hiv/intervention.py:139: RuntimeWarning:
Not enough candidates for ART. Expected: 110, Available: 0
ss.warn(msg=f"Not enough candidates for ART. Expected: {delta}, Available: {len(candidates)}")
/var/folders/dr/x377cvd10rl0xw0c1tj1vvzh0000gn/T/ipykernel_80111/1139370148.py:10: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
cmap = plt.cm.get_cmap('tab10', len(flat_results))