How to create a TB simulation¶
High level steps¶
As suggested by the starsim ‘Getting Started’ tutorial (https://docs.idmod.org/projects/starsim/en/latest/tutorials/tut_intro.html) we will proceed with the most common tasks for modeling infectious diseases, in our case, TB:
Defining parameters (Building the simulation):
Running a simulation
Plotting results
This notebook demonstrates how to create and run simulations for tuberculosis (TB) using the tbsim and starsim libraries. The simulations will model the spread and impact of TB over time on a population.
Packages¶
The code in this section imports the starsim package, which provides the capabilities to create and run simulations. Note that while tbsim enables some functionality, the full capabilities are provided by starsim.
[1]:
# Import necessary libraries
import tbsim as mtb
import starsim as ss
import matplotlib.pyplot as plt
import numpy as np
Building the simulation¶
In this case, in order to make our code reusable we will encapsulate all our preparation steps in the following function, make_tb. This function puts together all what is needed for a simulation to run and returns a simulation object with a tuberculosis disease, methods, etc., and with user specified parameters. NOTE: Default values are generally specified as part of the init method for each class, i.e. people, network, tb, etc.).
People:¶
Creates a population of 1,000 agents using the People class from the starsim (aliased as ss) library.
TB Disease¶
Defines the disease parameters for TB:
beta: Transmission rate (default: 0.025 per year using ss.peryear()).
init_prev: Initial prevalence of TB (default: 10% using ss.bernoulli()).
Initializes the TB model with these parameters using the tbsim library (aliased as mtb).
The TB class includes comprehensive disease progression with multiple states: latent (slow/fast), active (pre-symptomatic, smear positive/negative, extra-pulmonary), and treatment effects.
Network¶
Defines the network parameters:
n_contacts: Number of contacts per agent, following a Poisson distribution with a mean (lambda) of 5.
dur: Duration of contact (0 means end after one timestep).
Initializes a random network with these parameters.
Demographics¶
The demographics section sets up two demographic processes for the simulation: pregnancy and death rates. These, define and incorporate key demographic events into the simulation model:
Pregnancy: Adds new agents to the population at a rate of 25 per 1,000 people. (TODO: Verify if this is done every time every step or if this is a one time event at set_prognoses)
Deaths: Removes agents from the population at a rate of 10 per 1,000 people. (same question as above)
This setup ensures the simulation realistically accounts for population changes over time due to births and deaths.
Simulation Parameters and Initialization¶
This part of the code defines the parameters for the simulation and initializes it using these parameters:
Defining Simulation Parameters:
``dt = ss.days(7)``: This defines the time step (
dt) for the simulation. Here,ss.days(7)represents a weekly time step, so the simulation progresses in weekly increments.``start = ss.date(1990,1,1)``: This sets the starting date of the simulation to January 1, 1990.
``stop = ss.date(2020,1,1)``: This sets the ending date of the simulation to January 1, 2020.
Initializing the Simulation:
``ss.Sim``: This is a class from the
starsimpackage used to create a simulation object.Parameters:
``people=pop``: Specifies the population object created earlier.
``networks=net``: Specifies the network object that describes how agents are connected.
``diseases=tb``: Specifies the TB disease model initialized earlier.
``pars=sim_pars``: Passes the simulation parameters defined above.
``demographics=dems``: Specifies the list of demographic events (pregnancy and death rates).
Setting Verbosity:
This line adjusts how frequently the simulation prints its status updates.
``sim.pars.verbose = 0``: This sets the verbosity to 0, which means no status updates will be printed during the simulation run.
Returning the Simulation Object:
This returns the initialized simulation object so that it can be run or further interacted with outside of the
make_tbfunction.
[2]:
# Define the function to create a tuberculosis simulation
def make_tb():
# --------------- People ----------
initial_population_size = 1000
pop = ss.People(n_agents=initial_population_size)
# --------------- TB disease --------
# Using current default parameters from TB class
tb_pars = dict( # Disease parameters
beta = ss.peryear(0.025), # Updated to match current default
init_prev = ss.bernoulli(0.10), # 10% initial prevalence
)
tb = mtb.TB(tb_pars) # Initialize
# --------------- Network ---------
net_pars = dict( # Network parameters
n_contacts=ss.poisson(lam=5),
dur = 0, # End after one timestep
)
net = ss.RandomNet(net_pars) # Initialize a random network
# --------------- Demographics --------
dems = [
ss.Pregnancy(pars=dict(fertility_rate=25)), # Per 1,000 people
ss.Deaths(pars=dict(death_rate=10)), # Per 1,000 people
]
# --------------- simulation -------
sim_pars= dict(
dt = ss.days(7),
start = ss.date(1990,1,1),
stop = ss.date(2020,1,1),
)
sim = ss.Sim(people=pop,
networks=net,
diseases=tb,
pars=sim_pars,
demographics=dems
) # initialize the simulation
sim.pars.verbose = 0 # Print status every 100 steps
return sim
Run the Simulation¶
We will now create and run the first simulation using the make_tb function with the parameters set above.
Please note, at some point it may be convenient to parameterize the make_tb function and make it even more re-usable (for intance for scenarios creation).
[3]:
# Create and run the first simulation
sim = make_tb() # Create the simulation - running make_tb returns a simulation of a type ss.Sim (from starsim library) therefore we can use all the methods of the ss.Sim class
sim.run() # Run the simulation
[3]:
Sim(n=1000; 1990.01.01—2020.01.01; demographics=pregnancy, deaths; networks=randomnet; diseases=tb)
Analyzing The Results¶
Let’s first review the data that was generated (and is available in the results array). This collected data is specified in the init_results() method and then updated in the update_results() method of the TB class. The results include:
State counts: Number of individuals in each TB state (latent, active, infectious)
Age-specific metrics: Separate tracking for individuals 15+ years old
Incidence and mortality: New cases, deaths, and cumulative measures
Epidemiological indicators: Prevalence, incidence rates, and detection metrics
For more detailed information, please refer to the official documentation at StarSim Results API.
[4]:
tbr = sim.diseases['tb'].results
print("This is a dictionary of the results that you can plot: \n",tbr.keys())
This is a dictionary of the results that you can plot:
['timevec', 'n_susceptible', 'n_infected', 'n_on_treatment', 'n_ever_infected', 'prevalence', 'new_infections', 'cum_infections', 'n_latent_slow', 'n_latent_fast', 'n_active', 'n_active_presymp', 'n_active_presymp_15+', 'n_active_smpos', 'n_active_smpos_15+', 'n_active_smneg', 'n_active_smneg_15+', 'n_active_exptb', 'n_active_exptb_15+', 'new_active', 'new_active_15+', 'cum_active', 'cum_active_15+', 'new_deaths', 'new_deaths_15+', 'cum_deaths', 'cum_deaths_15+', 'n_infectious', 'n_infectious_15+', 'prevalence_active', 'incidence_kpy', 'deaths_ppy', 'n_reinfected', 'new_notifications_15+', 'n_detectable_15+']
now, below we show the plotting of a single channel (result):
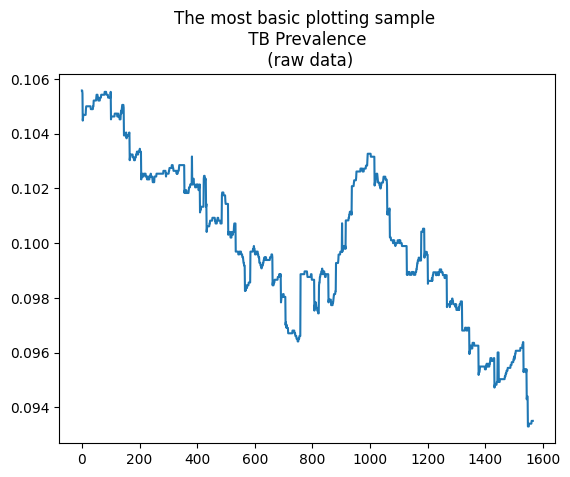
[5]:
plt.plot(range(0,len(tbr['prevalence'])),tbr['prevalence'])
plt.title("The most basic plotting sample \n TB Prevalence \n (raw data)")
[5]:
Text(0.5, 1.0, 'The most basic plotting sample \n TB Prevalence \n (raw data)')
##
Plotting
Once you get familiar with the available data in the results array, you can proceed to plot your results using your favorite method.
###
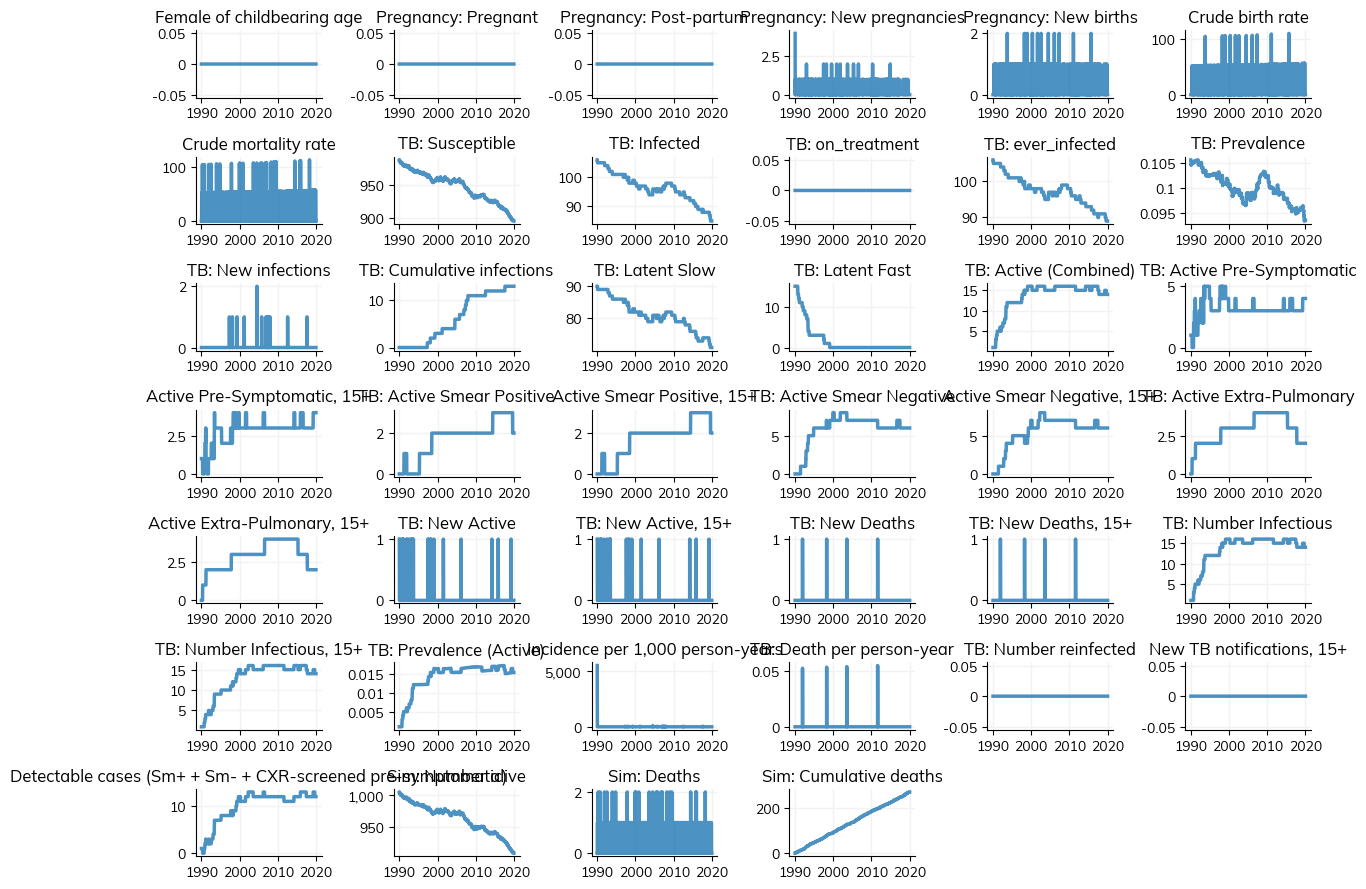
1: Using the starsim library
Plot the results of the simulation - inherited from the starsim library which is available under the diseases attribute of the simulation: sim.plot()
[6]:
sim.plot()
Figure(1200x900)
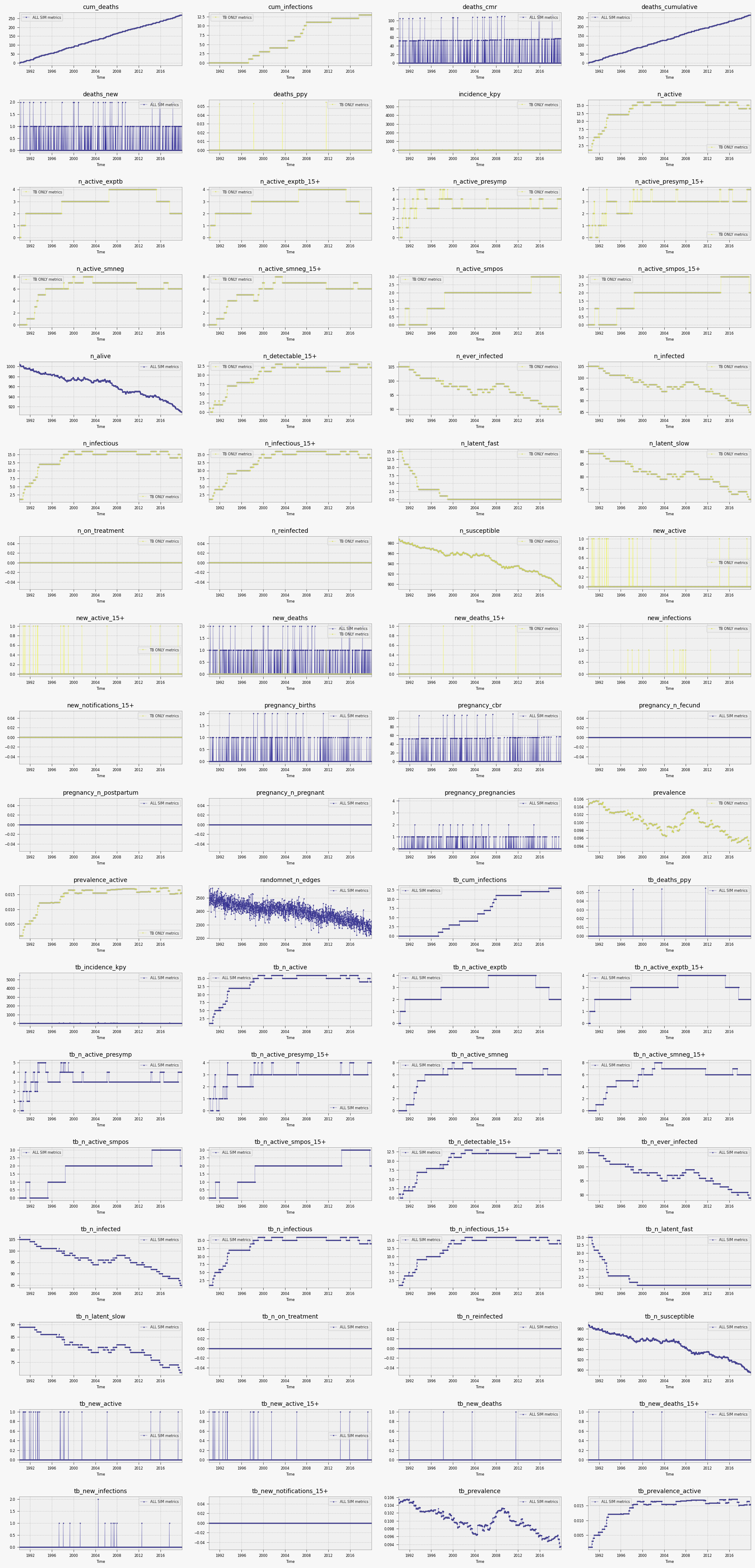
Using TBSim plots:¶
The plot_combined function from tbsim can be used to create comprehensive plots:
mtb.plot_combined({'single simulation': sim.results.flatten(), 'tb only results': sim.diseases['tb'].results.flatten()})
where each key represents a scenario that will be plotted with a different color.
For more information, see the plot_combined function documentation.
[7]:
# Create comprehensive plots using tbsim's plot_combined function
mtb.plot_combined({'ALL SIM metrics': sim.results.flatten(),'TB ONLY metrics': sim.diseases['tb'].results.flatten()}, dark=False, n_cols=4, heightfold=2 )
Saved figure to /Users/mine/newgit/newtbsim/docs/tutorials/results/scenarios_20250907_222603.png
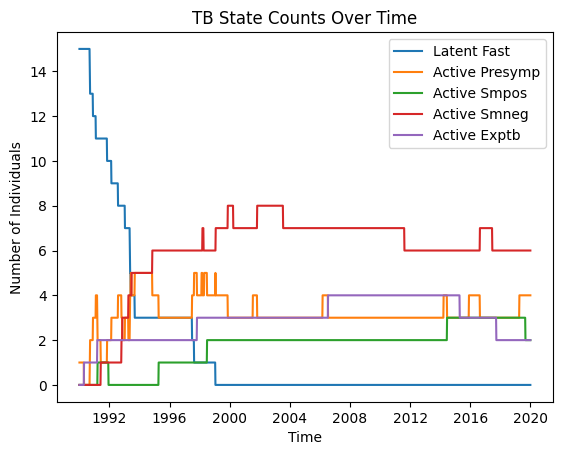
Using your own function¶
Take a look at the sample below, it will show you how to access the results dataframe (dict) and how with a simple function you can plot your simulation’s results.
[8]:
# Method 3: Custom plotting using TB results directly
fig = plt.figure()
tb_results = sim.diseases['tb'].results
for rkey in ['n_latent_fast', 'n_active_presymp', 'n_active_smpos', 'n_active_smneg', 'n_active_exptb']:
plt.plot(tb_results['timevec'], tb_results[rkey], label=rkey.replace('n_', '').replace('_', ' ').title())
plt.legend()
plt.title("TB State Counts Over Time")
plt.xlabel("Time")
plt.ylabel("Number of Individuals")
[8]:
Text(0, 0.5, 'Number of Individuals')
Additional Information¶
Current TB Model Features¶
The TB model in this notebook uses the latest implementation from the tbsim library, which includes:
Comprehensive Disease States: The model tracks multiple TB states including latent (slow/fast progression), active (pre-symptomatic, smear positive/negative, extra-pulmonary), and treatment effects.
Realistic Parameters: Default parameters are based on epidemiological literature and include:
Transmission rate: 0.025 per year
Initial prevalence: 10% of population (as used in this tutorial)
State-specific progression rates and mortality
Treatment effects on transmission and mortality
Age-Specific Tracking: Separate metrics for individuals 15+ years old, important for TB epidemiology.
Treatment Integration: The model includes treatment effects that reduce transmission and prevent mortality.
Next Steps¶
To extend this simulation, you can:
Add interventions (diagnostics, treatment programs, vaccination)
Include comorbidities (HIV, malnutrition)
Modify network structures (household networks, spatial networks)
Run scenario analyses with different parameter sets
Use the comprehensive analyzers for detailed epidemiological analysis
For more advanced examples, see the other tutorials in this documentation.