HIV Comorbidity in TBSim¶
This notebook demonstrates how to use the HIV comorbidity module in the tbsim package.
Module Location: tbsim/comorbidities/hiv/
Overview¶
The HIV comorbidity module in TBSim allows users to simulate the impact of HIV infection on TB disease dynamics. Rather than modeling HIV progression in detail, it serves as a lightweight comorbidity that modifies TB risks based on HIV state.
The module is composed of three key components:
HIV Model: Represents HIV infection as a comorbidity.
HIV Intervention: Enables scenario testing via prevalence and ART coverage.
TB-HIV Connector: Bridges HIV and TB models, applying comorbidity risk logic.
HIV Model¶
The HIV model simulates HIV infection status and its progression through simplified states:
ACUTELATENTAIDS
This implementation is intended as a modifier for TB outcomes, rather than a standalone disease model.
Key Parameters¶
Parameter |
Description |
|---|---|
|
Probability of being HIV-positive (ACUTE stage) at simulation start |
|
Probability of being on ART if HIV-positive |
|
Daily probability of transitioning from ACUTE to LATENT |
|
Daily probability of transitioning from LATENT to AIDS |
|
Daily probability of death after reaching AIDS (currently unused) |
|
Multiplier reducing HIV progression for agents on ART |
*** Important: ***¶
At step 0, the model looks for the existance of an
HivInterventionhandling the HIV infection and art coverage, if none is found then it uses the values defined at Disease level to set the initial values.
HIV Intervention¶
The HivIntervention class allows control over HIV prevalence and ART coverage in the population. This is useful for scenario testing and simulating public health campaigns.
Supported Modes¶
'infection': Adjust HIV infection prevalence'art': Adjust ART coverage'both': Adjust both infection and treatment
What It Does¶
Applies changes to the HIV state of the population
Operates within a specified time range (
starttostop)Can be targeted to specific age groups or entire population
intervention = HivInterventions(pars=dict(
mode='both',
prevalence=0.2,
percent_on_ART=0.5,
minimum_age=15,
max_age=49,
start=ss.date('2000-01-01'),
stop=ss.date('2010-12-31'),
))
Also, the parameters shown above are the same ones that you can modify in order to run a different scenario.
TB-HIV Connector¶
The TB_HIV_Connector class links the TB and HIV models, simulating the comorbidity effect of HIV on TB progression.
Purpose¶
Modifies TB progression parameters based on an individual’s current HIV state.
Risk Multiplier Table¶
HIV State |
TB Progression Risk Multiplier |
|---|---|
|
1.5 |
|
2.0 |
|
3.0 |
Behavior at Each Time Step¶
Identify all TB-infected individuals.
Look up their current HIV state.
Apply a multiplier to their TB
rr_activationvalue (risk of progressing to active TB).
Process Flow Diagram¶
+-----------------+
| TB Infection |
+-----------------+
|
v
+-----------------+ +-----------------------+
| HIV Comorbidity| --> | HIV State (ACUTE,...) |
+-----------------+ +-----------------------+
| |
| TB_HIV_Connector |
+-------------------------> Applies RR Multiplier
|
v
+-------------------------------+
| TB rr_activation Adjusted |
| Based on HIV State |
+-------------------------------+
Example Code¶
Example¶
[64]:
import tbsim as mtb
import starsim as ss
import sciris as sc
import numpy as np
import matplotlib.pyplot as plt# Rendering with Quatro
Intervention¶
Adding a function to simplify the generation of an intervention.
This intervention will try to simulate a steady prevalence and on ART values along the entire simulation
[65]:
def make_hiv_intervention(pars=None):
if pars is None:
pars=dict(
mode='both',
prevalence=0.30, # Maintain 30 percent of the alive population infected
percent_on_ART=0.50, # Maintain 50 percent of the % infected population on ART
min_age=15, max_age=60, # Min and Max age of agents that can be hit with the intervention
start=ss.date('2000-01-01'), stop=ss.date('2035-12-31'), # Intervention's start and stop dates
)
intervention = mtb.HivInterventions(pars=pars)
return intervention
Creating the simulation¶
[66]:
def build_tbhiv_sim(simpars=None, tbpars=None, hivinv_pars=None) -> ss.Sim:
sim_pars = dict(
dt=ss.days(7),
start=ss.date('1980-01-01'),
stop=ss.date('2035-12-31'),
rand_seed=123,
verbose=0,
)
tb_pars = dict(
init_prev=ss.bernoulli(p=0.25),
rel_sus_latentslow=0.1,
)
hivinv_pars = hivinv_pars or dict(
mode='both',
prevalence=0.20,
percent_on_ART=0.20,
min_age=15, max_age=49,
start=ss.date('2000-01-01'),
stop=ss.date('2010-12-31'),
)
hiv_intervention = make_hiv_intervention(hivinv_pars)
hiv = mtb.HIV()
connector = mtb.TB_HIV_Connector()
tb = mtb.TB(pars=tb_pars)
people = ss.People(n_agents=1_000)
network = ss.RandomNet(pars=dict(n_contacts=ss.poisson(lam=2), dur=0))
# --- Assemble Simulation ---
sim = ss.Sim(
people=people,
diseases=[tb, hiv],
interventions=[hiv_intervention],
networks=network,
connectors=[connector],
pars=sim_pars,
)
return sim
Running the simulation¶
[67]:
# Run the simulation:
sim = build_tbhiv_sim()
sim.run()
HIV intervention present, skipping initialization.
[67]:
Sim(n=1000; 1980.01.01—2035.12.31; connectors=tb_hiv_connector; networks=randomnet; interventions=hivinterventions; diseases=tb, hiv)
Plot the results¶
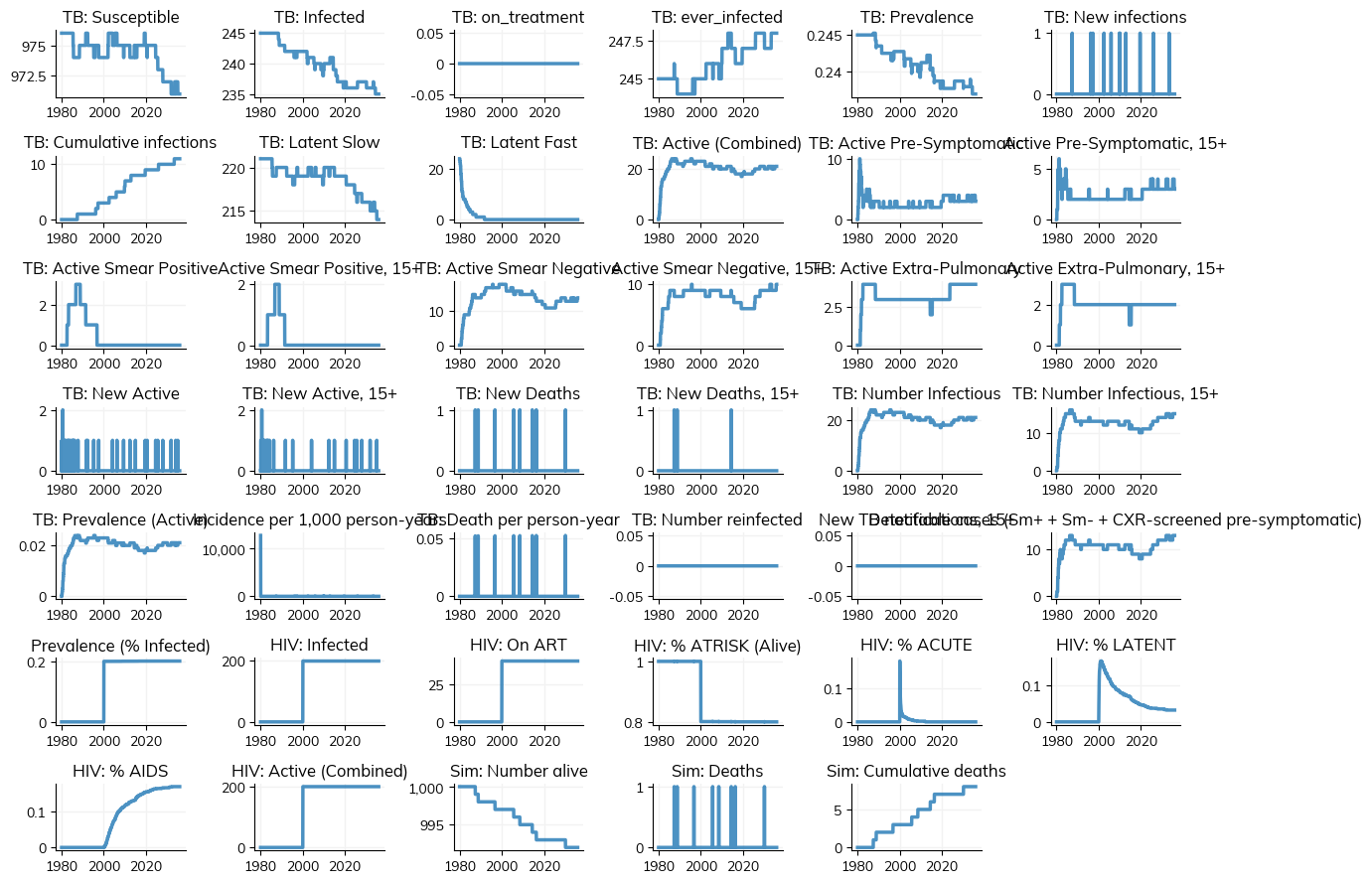
[68]:
sim.plot()
plt.show()
Figure(1200x900)